Gene Expression Data Analysis Suite (GEDAS)
Tree view
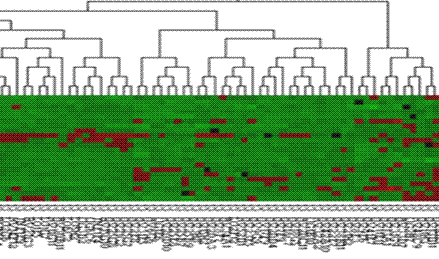
· Description or interpretation: In HC, the tree view (dendrogram) output is the most common output; however, in an attempt to standardize and provide common outputs to all the data mining applications, we were successful in converting the tree view output into cluster (or temporal) view using our GEDAS software. This extension of outputs enabled further visualization using other methods such as microarray view, textual view, whole genome view, etc. One of the most effective and powerful representation of clustered gene expression data is the tree view or dendrogram view consisting of three portions viz., the gene tree, array tree and the colour coded band of gene expression, see following figure. This representation is sometimes also known as matrix tree plots or two way dendrograms.
· Complexity: While the computational complexity of the distance matrix is not included, two different algorithms work together to build up the gene tree on one side and array tree and the colour coded band of expression values on the other side.
· Special considerations/features: The user can be given control to select the colour of representing low to high gene expression such as green to red or blue to red or so on. Different colour codes can be assigned to represent null values or zero values. Shades represent the intensity or magnitude of expression.
·
Advantages and drawbacks: It offers clustering of both the
genes and samples simultaneously. However, if the dataset is very large,
it also requires another GUI support to extract the gene names mined out.
It is very helpful for studying the trend in time series data and data of same
parameter over different samples.
|
|
|
|
|
Figure: The dendrogram or tree view
can be constructed for genes as well as samples |